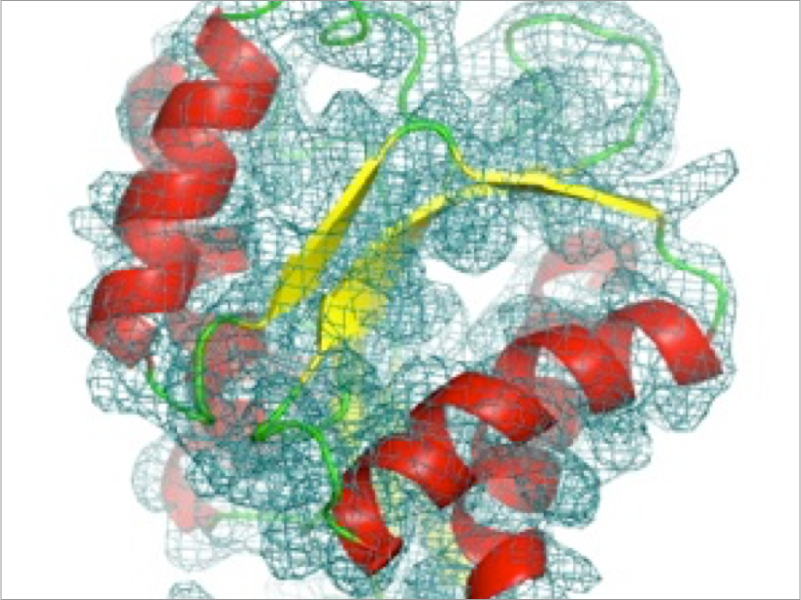
1. Protein structure modeling based on the cryo-EM data
Single-particle cryo-EM is a powerful tool to determine three-dimensional structures of biomolecules. Since the method does not require crystallization of the sample, structures of large protein complexes can be determined near atomic resolution, which had been difficult so far with X-ray crystallography. To construct the atomic structure of the protein complex from its cryo-EM density map, it is useful to utilize a flexible fitting method, where the component proteins determined with X-ray crystallography or NMR are fitted to the map using MD techniques. We are developing new methods for the cryo-EM flexible fitting using our MD program GENESIS towards reliable structure modeling of large protein complexes.
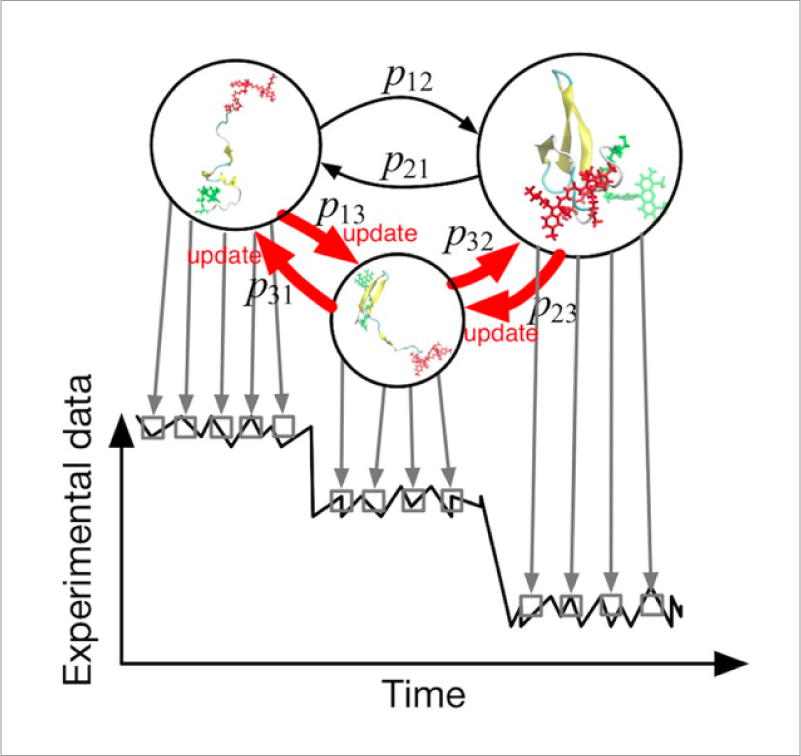
2. Towards understating of protein dynamics using data science
Large structural changes of biomolecules are strongly related to cellular functions such as enzymatic reaction and signal transduction. To understand their molecular mechanisms, it is essential to analyze dynamic structures of proteins at high resolution. Although MD simulations can provide information about protein dynamics at the atomic resolution, the simulations are not always reliable due to incompleteness of the model parameters. Single-molecule FRET experiments can provide information about structural dynamics. However, it is still difficult to understand detailed structural changes of proteins only from distance data. We are developing new methods to combine high-resolution simulation model and low-resolution experimental data based on the data science such as machine learning and data assimilation to understand protein dynamics.
References
- “Acceleration of cryo-EM Flexible Fitting for Large Biomolecular Systems by Efficient Space Partitioning”, Takaharu Mori, Marta Kulik, Osamu Miyashita, Jaewoon Jung, Florence Tama and Yuji Sugita, Structure, 27, 161-174 (2019).
- “Flexible Fitting to Cryo-EM Density Map Using Ensemble Molecular Dynamics Simulations”, Osamu Miyashita, Chigusa Kobayashi, Takaharu Mori, Yuji Sugita, and Florence Tama., J. Comp. Chem., 38, 1447-1461 (2017).
- “Linking time-series of single-molecule experiments with molecular dynamics simulations by machine learning”, Yasuhiro Matsunaga and Yuji Sugita, eLife, 7, e32668 (2018).
- “Sequential data assimilation for single-molecule FRET photon-counting data”, Yasuhiro Matsunaga, Akinori Kidera, and Yuji Sugita., J. Chem. Phys., 142, 214115 (2015).
- “Characterization of Conformational Ensembles of Protonated N-glycans in the Gas-Phase”, Suyong Re, Shigehisa Watabe, Wataru Nishima, Eiro Muneyuki, Yoshiki Yamaguchi, Alexander D. MacKerell Jr. and Yuji Sugita, Scientific Reports, 8, 1644 (2018).
Collaborators
Osamu Miyashita (RIKEN R-CCS), Florence Tama (RIKEN R-CCS, Nagoya Univ.), Yasuhiro Matsunaga (Saitama Univ.), Yasushi Sako (RIKEN CPR)