Research Projects
Project 1: Spectral Analysis and Reaction Mechanism by QM/MM Calculations
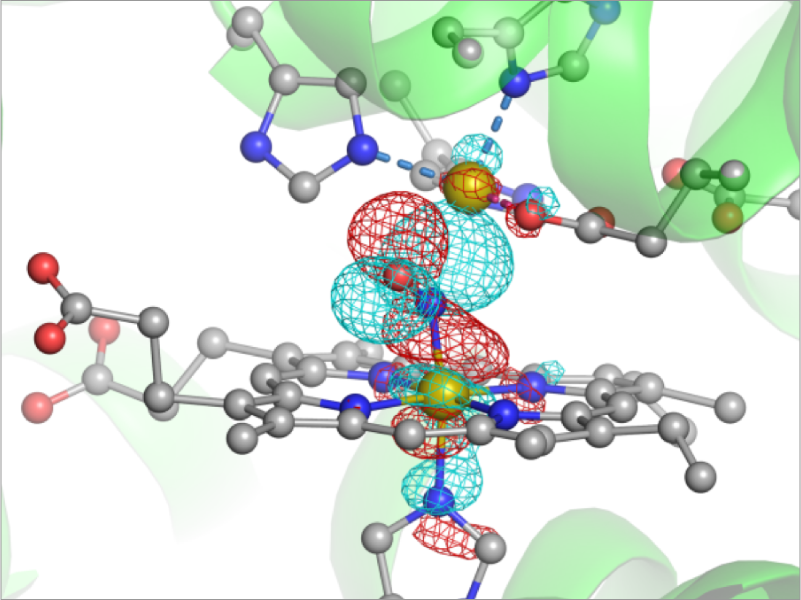
The understanding of chemical reactions in biomolecules, such as proton transfers and enzymatic reactions, requires a theoretical treatment that can describe bond breaking and forming processes. We develop a hybrid QM/MM approach, which describes the reaction center by quantum chemical (QM) methods and the surrounding environment by the molecular mechanics (MM) force field. We utilize the method to search the reaction path and reveal the mechanism of enzymatic reactions. We also calculate various spectra such as Infrared/Raman, UV-Vis, etc. in collaboration with experimental spectroscopists. The goal of this project is to understand the mechanism of enzymatic reactions in cellular environment .
Project 2: Data-driven simulations to understand protein dynamics
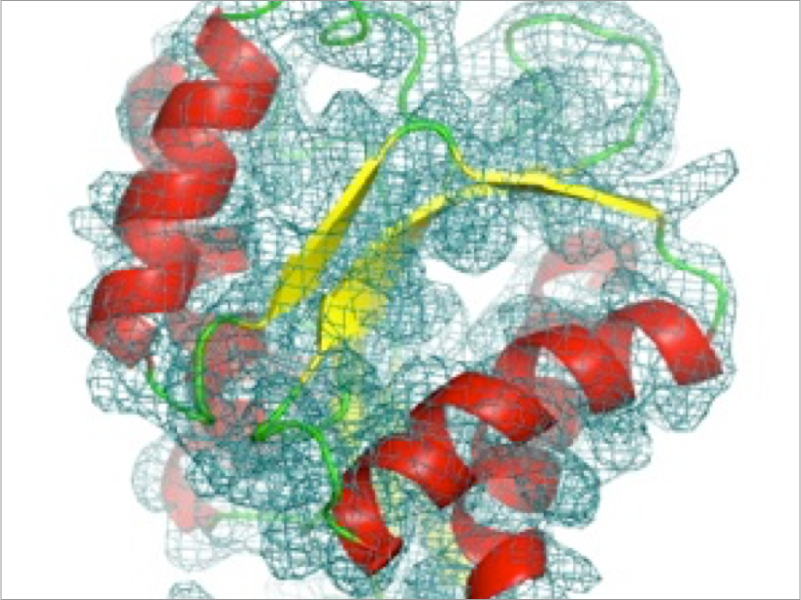
Recently, experimental methods such as X-ray crystallography, NMR, cryo-EM, High-speed AFM, and single molecule FRET have been improved to obtain detailed structure information of proteins at high resolution, enabling us to compare directly with MD simulations. We are developing new methods to combine experimental data and MD simulations towards reliable understanding of protein structure-dynamics-function relationships.
Project 3: Molecular crowding in the cell
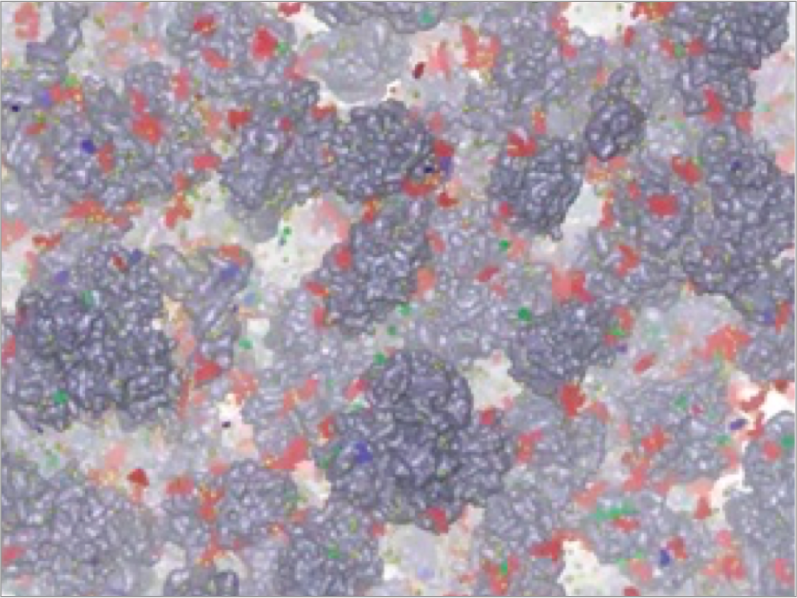
The cell is mainly composed of water. However, the cytoplasm is a kind of “crowded environments”, where the macromolecules such as proteins and RNAs are highly concentrated. Detailed molecular mechanisms of enzymatic reactions or protein networks formation in such a crowded environment have not been understood well. We are attempting to analyze biophysical properties of macromolecular crowding by performing cellular scale MD simulations using supercomputers, and also by collaborating with experimental groups of in-cell NMR.
Gallery
Movies and figures in our research activity
Facilities
Research facilities in our laboratory