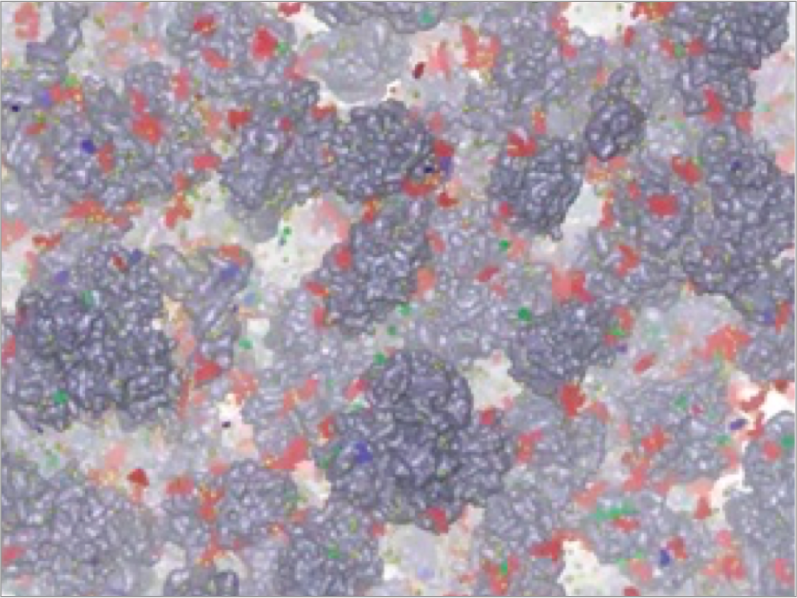
The cell is mainly composed water, and the remaining are proteins, nucleic acids, metabolites, and ions. Proteins are highly concentrated in the cell, which is called “macromolecular crowding”. Understanding of structures, dynamics, and functions of proteins in the cell at the molecular level is one of the challenging issues both experimentally and theoretically. Recent in-cell NMR experiments have suggested that some proteins are stabilized in the cellular environment compared to the dilute solution, while some are destabilized. We are trying to construct a detailed model of macromolecular crowding of the cytoplasmic system, and analyze protein dynamics using all-atom MD simulations with GENESIS and supercomputers such as K computer.
References
- “Slow-Down in Diffusion in Crowded Protein Solutions Correlates with Transient Cluster Formation”, Grzegorz Nawrocki, Po-hung Wang, Isseki Yu, Yuji Sugita, and Michael Feig, J. Phys. Chem. B, 121, 11072–11084 (2017).
- “Crowding in Cellular Environments at an Atomistic Level from Computer Simulations”, Michael Feig, Isseki Yu, Po-hung Wang, Grzegorz Nawrocki, and Yuji Sugita, J. Phys. Chem. B, 121, 8009-8025 (2017).
- “Influence of protein crowder size on hydration structure and dynamics in macromolecular crowding”, Po-hung Wang, Isseki Yu, Michael Feig, and Yuji Sugita., Chem. Phys. Lett., 671, 63-70 (2017).
- “Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm”, Isseki Yu, Takaharu Mori, Tadashi Ando, Ryuhei Harada, Jaewoon Jung, Yuji Sugita, and Michael Feig., eLife, 5, e19274 (2016).
- “Thermodynamics of Macromolecular Association in Heterogeneous Crowding Environments: Theoretical and Simulation Studies with a Simplified Model”, Tadashi Ando, Isseki Yu, Michael Feig, and Yuji Sugita., J. Phys. Chem. B, 120, 11856-11865 (2016).
- “Complete Atomistic Model of a Bacterial Cytoplasm for Integrating Physics, Biochemistry, and Systems Biology Journal of Molecular Graphics and Modelling”, Michael Feig, Ryuhei Harada, Takaharu Mori, Isseki Yu, Koichi Takahashi and Yuji Sugita., J. Mol. Graph. Model., 58, 1–9 (2015).
- “Reaching new levels of realism in modeling biological macromolecules in cellular environments”, Michael Feig and Yuji Sugita., J. Mol. Graph. Model., 45, 144–156 (2013).
- “Reduced Native State Stability in Crowded Cellular Environment Due to Protein–Protein Interactions”, Ryuhei Harada, Naoya Tochio, Takanori Kigawa, Yuji Sugita and Michael Feig., J. Am. Chem. Soc., 135, 3696–3701 (2013).
- “Protein Crowding Affects Hydration Structure and Dynamics”, Ryuhei Harada, Yuji Sugita and Michael Feig., J. Am. Chem. Soc., 134, 4842-4849 (2012).
Collaborators
Michael Feig (Michigan State University), Takanori Kigawa (RIKEN BDR), Yutaka Ito (Tokyo Metropolitan Univ.), Isseki Yu (Maebashi Institute of Technology)