1. Development of QM/MM method in GENESIS
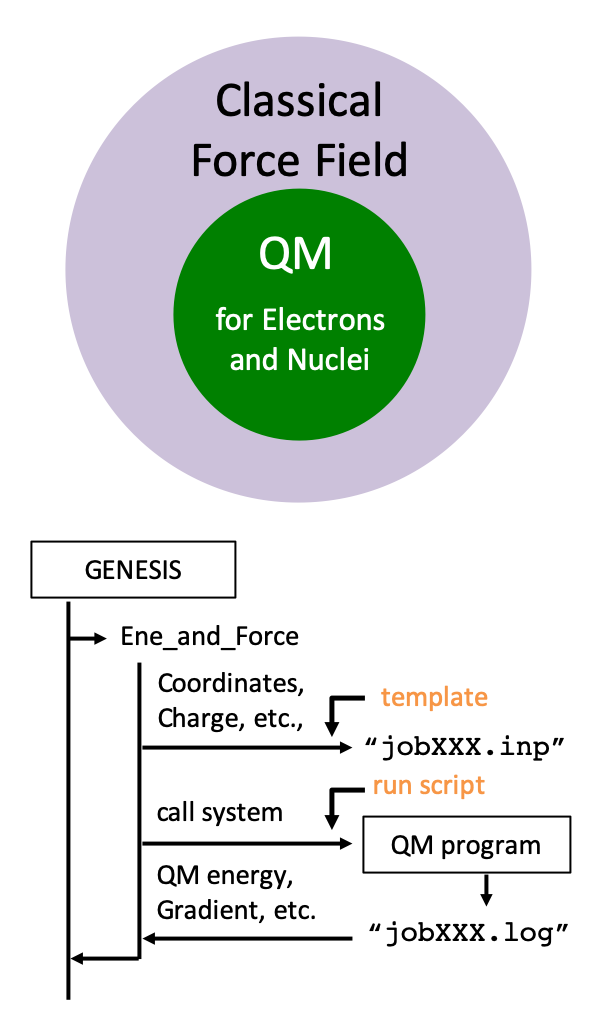
QM/MM method is a hybrid approach, where a local region of interest is treated by quantum chemistry (QM) calculations and the surrounding environment by molecular mechanics (MM) force fields. The method makes it feasible to calculate molecular properties of complex systems, such as spectra, chemical reaction paths, etc., with reasonable cost. We have recently implemented the QM/MM method into GENESIS. GENESIS has an interface with QM programs to carry out QM/MM calculations. In the energy routine, GENESIS generates an input file for a QM program, execute the program, and reads from output files the QM energy, gradient, etc. Currently, the interface with Gaussian, Q-Chem, DFTB+, and TeraChem is available. We aim at developing an efficient method for QM/MM-MD simulations, which incorporates both sufficient structural samplings and accurate electronic structures.
2. Vibrational analyses of biomolecules
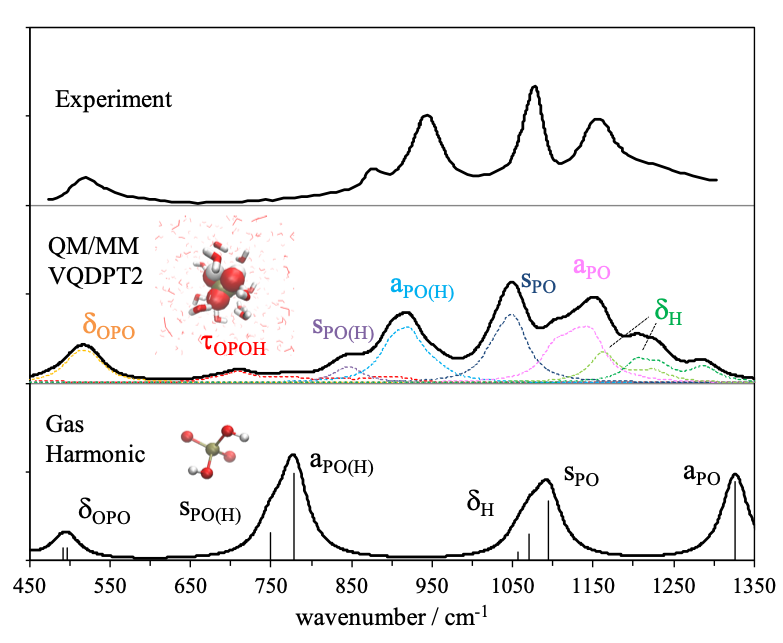
Hydrogen bonds play an important role in biomolecules. For example, the three-dimensional structure of proteins is formed by hydrogen bonds between amino acids, and proton transfer reactions are often mediated by strong hydrogen bonds. Vibrational spectroscopy is a viable tool to detect hydrogen bonds. Although theoretical calculations have been used to interpret the spectrum, the conventional harmonic approximation is insufficient in accuracy to compute the spectrum of OH/NH stretching vibration in the presence of hydrogen bonds. We have developed an efficient solver of vibrational Schrödinger equation, anharmonic vibrational analysis method based on QM/MM, and a weight average approach utilizing structural sampling by MD simulations. The developed methods are applied to biomolecules.
3. The reaction mechanism of enzymatic reaction
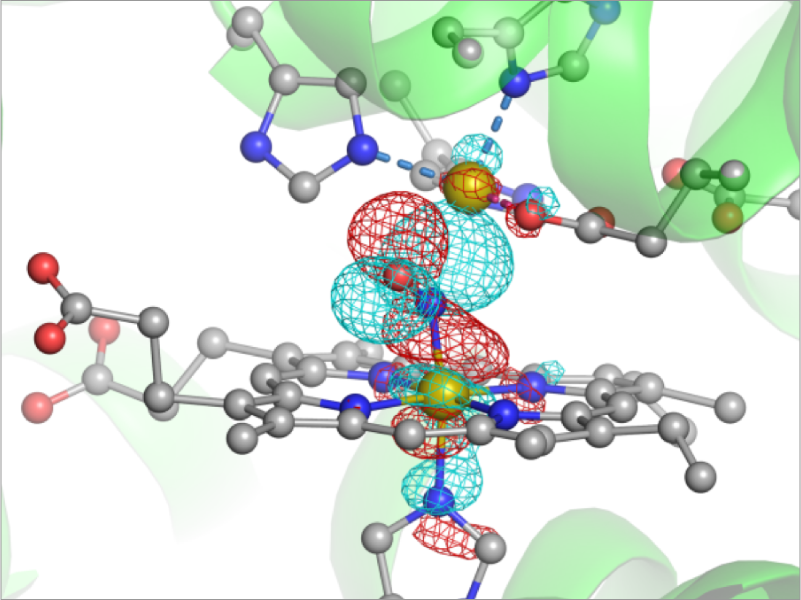
A number of chemical reactions take place in biological environment. For example, oxygen reduction reaction is the source of energy for biological activities. The phosphorylation and de-phosphorylation of proteins are key processes in cell signaling to adjust the activity of protein functions. Enzymes catalyze these reactions. Therefore, understanding the molecular mechanism of enzyme reactions is of great importance. We develop a method to compute the reaction path based on QM/MM. The developed method is applied to nitric oxide reductase (NOR). NOR is an enzyme that reduces NO molecules to N2O, and vital for anaerobic bacteria to produce the energy. We have revealed using the X-ray crystallography and MD simulations that nitrite oxide reductase (NiR), which produces NO by reducing NO2– , forms a complex with NOR so as to prevent toxic NO from diffusion in cell. On the other hand, the reaction of NO reduction in NOR is still controversial. We employ QM/MM method to study the details.
References
- “Anharmonic Vibrational Analysis of Biomolecules and Solvated Molecules Using Hybrid QM/MM Computations.” Kiyoshi Yagi, Kenta Yamada, Chigusa Kobayashi, and Yuji Sugita, J. Chem. Theory Comput, 3, 1924-1938 (2019).
- “Dynamics of nitric oxide controlled by protein complex in bacterial system”, Erina Terasaka, Kenta Yamada, Po-Hung Wang, Kanta Hosokawa, Raika Yamagiwa, Kimi Matsumoto, Shoko Ishii, Takaharu Mori, Kiyoshi Yagi, Hitomi Sawai, Hiroyuki Arai, Hiroshi Sugimoto, Yuji Sugita, Yoshitsugu Shiro, and Takehiko Tosha., Proc. Natl. Acad. Sci. USA, 114, 9888–9893 (2017).
- “Weight-Averaged Anharmonic Vibrational Analysis of Hydration Structures of Polyamide 6”, Bo Thomsen, Tomonori Kawakami, Isamu Shigemoto, Yuji Sugita, and Kiyoshi Yagi., J. Phys. Chem. B, 121, 6050-6063 (2017).
- “Anharmonic Vibrational Analyses of Pentapeptide Conformations Explored with Enhanced Sampling Simulations”, Hiroki Otaki, Kiyoshi Yagi, Shun-ichi Ishiuchi, Masaaki Fujii, and Yuji Sugita., J. Phys. Chem. B, 120, 10199-10213 (2016).
- “A weight averaged approach for predicting amide vibrational bands of a sphingomyelin bilayer” Kiyoshi Yagi, Pai-Chi Li, Koichiro Shirota, Toshihide Kobayashi and Yuji Sugita, Phys. Chem. Chem. Phys., 17, 29113-29123 (2015).
Collaborators
Yoshitsugu Shiro (University of Hyogo)